Remote Sensing Zooming in from above.
Remote sensing data are critical for scaling up local observations of biodiversity, and for relating physical and ecological variables to marine biodiversity.
The SCB MBON is focused upon three activities linking remote sensing to observations of marine biodiversity:
- Remote sensing of giant kelp populations and the application of these products to answer ecological and biodiversity questions
- Analysis of satellite ocean color observations using novel ocean color inversion approaches
- Analysis of planktonic biodiversity indices from the Plumes and Blooms time-series study in parallel with SCB MBON genomics observations
Giant Kelp Populations
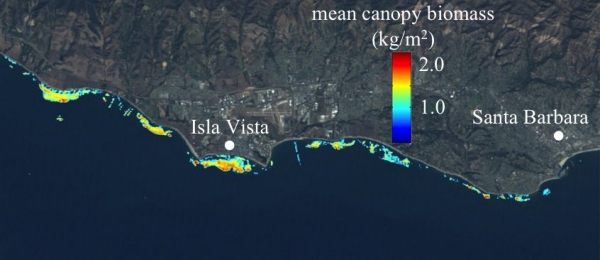
Both multispectral and hyperspectral remote sensing of giant kelp canopy fronds are used to help answer ecological and biodiversity questions for the SCB MBON. Led by Tom Bell and in collaboration with the Santa Barbara Coastal LTER, we have assembled a Landsat giant kelp (Macrocystis) canopy biomass time series from 1984 – present, spanning its dominant range in the NE Pacific (San Francisco, CA to Punta Eugenia, Baja California Sur, Mexico). This dataset has recently been used to determine the geographical variability and non-linear response of the environmental controls of giant kelp biomass dynamics (Bell et al. 2015a), as well as a test of metapopulation theory, showing that well connected patches had high probabilities of colonization and lower probabilities of extinction (Castorani et al. 2015). Presently, this dataset is being used to identify large-scale biogeographic population clusters using seascape genetic approaches (Johansson et al. in review). Multiple hyperspectral images are being collected as part of the HyspIRI Preparatory Airborne Campaign. Empirical relationships between laboratory reflectance of giant kelp blades and their physiological state (Chl:C) have been developed and applied to hyperspectral images captured by the AVIRIS sensor. These images have shown that the physiological state of the canopy is positively correlated to the depth of the reef where the kelp plants attach, and may have implications for measuring the productivity and age structure of kelp forests (Figure 1; Bell et al. 2015b).
Ocean Color Analysis
More than 13 years of NASA satellite ocean color observations are used to characterize the controls of space-time variability of optical properties of the Southern California Bight (SCB). Specifically, we are looking to understand how changes in chlorophyll concentrations (chl) and particulate backscattering coefficients (bbp) co-vary over time and space. Level-2 spectral ocean reflectance data from SeaWIFS, MODIS and MERIS satellites are merged spectrally following procedures developed by Maritorena et al. (2010; RSE) using the GSM semi-analytical algorithm to simultaneously retrieve values of chlorophyll concentration and particulate backscattering coefficient at 443nm. Validation between retrieved parameters and Plumes and Blooms field data is encouraging. Results thus far show the importance of upwelling, relaxation and storm events in determining optical variability over time, and the role of episodic events in driving general patterns of distribution is apparent. Chlorophyll and backscatter seem very well correlated in offshore waters, while the relationship breaks down within ~15km from the coast. The role of re-suspension of materials, upwelling conditions and photoacclimation in affecting the predictability of the backscatter-chlorophyll relationship throughout the SCB will be addressed. The importance of the submesoscale components of particle variability for determining nearshore productivity will be assessed.
Plumes and Blooms
The goal of the Plumes and Blooms (PnB) study is to assess and model ocean color changes in a complex coastal site and is supported by the NASA Ocean Biology and Biogeochemistry program. PnB conducts monthly day-long cruises at 7 stations crossing the Santa Barbara Channel (~45 km long transect). At each station, measurements of ocean color spectra, inherent optical properties, phytoplankton pigment, dissolved and particulate carbon, and macronutrient concentrations, and particle size spectra are measured. Samples are also collected for particulate DNA for both prokaryotic and eukaryotic. Our eventual goal in the SCB MBON is to statistically relate bio-optical information of planktonic biodiversity from the PnB measurement suite (phytoplankton pigment concentrations, phytoplankton absorption spectra, etc.) with signatures from the SCB MBON genomic work.